
ConSciR provides tools for the analysis of cultural
heritage preventive conservation data.
It includes functions for environmental data analysis, humidity calculations, sustainability metrics, conservation risks, and data visualisations such as psychrometric charts. It is designed to support conservators, scientists, and engineers by streamlining common calculations and tasks encountered in heritage conservation workflows. The package is motivated by the framework outlined in Cosaert and Beltran et al. (2022) “Tools for the Analysis of Collection Environments” “Tools for the Analysis of Collection Environments”.
ConSciR is intended for:
- Conservators working in museums, galleries, and heritage sites
- Conservation scientists, engineers, and researchers
- Data scientists developing applications for conservation
- Cultural heritage professionals involved in preventive
conservation
- Students and educators in conservation and heritage science
programmes
The package is also designed to be:
- FAIR: Findable, Accessible, Interoperable, and
Reusable
- Collaborative: enabling contributions, feature
requests, bug reports, and additions from the wider community
If using R for the first time, read an article here: Using R for the first time
install.packages("ConSciR")
library(ConSciR)You can install the development version of the package from GitHub
using the pak package:
install.packages("pak")
pak::pak("BhavShah01/ConSciR")
# Alternatively
# install.packages("devtools")
# devtools::install_github("BhavShah01/ConSciR")For full details on all functions, see the package Reference manual.
This section demonstrates some common tasks you can perform with the ConSciR package.
library(ConSciR)
library(dplyr)
library(ggplot2)mydata) is included for testing and
demonstration. Use head() to view the first few rows and
inspect the data structure.mydata to ensure compatibility with ConSciR functions,
which expect variables such as temperature and relative humidity in
specific column names.# My TRH data
filepath <- data_file_path("mydata.xlsx")
mydata <- readxl::read_excel(filepath, sheet = "mydata")
mydata <- mydata |> filter(Sensor == "Room 1")
head(mydata)
#> # A tibble: 6 × 5
#> Site Sensor Date Temp RH
#> <chr> <chr> <dttm> <dbl> <dbl>
#> 1 London Room 1 2024-01-01 00:00:00 21.8 36.8
#> 2 London Room 1 2024-01-01 00:15:00 21.8 36.7
#> 3 London Room 1 2024-01-01 00:29:59 21.8 36.6
#> 4 London Room 1 2024-01-01 00:44:59 21.7 36.6
#> 5 London Room 1 2024-01-01 00:59:59 21.7 36.5
#> 6 London Room 1 2024-01-01 01:14:59 21.7 36.2# Peform calculations
head(mydata) |>
mutate(
# Dew point
DewP = calcDP(Temp, RH),
# Absolute humidity
Abs = calcAH(Temp, RH),
# Mould risk
Mould = ifelse(RH > calcMould_Zeng(Temp, RH), "Mould risk", "No mould"),
# Preservation Index, years to deterioration
PI = calcPI(Temp, RH),
# Scenario: Humidity if the temperature was 2°C higher
RH_if_2C_higher = calcRH_AH(Temp + 2, Abs)
) |>
glimpse()
#> Rows: 6
#> Columns: 10
#> $ Site <chr> "London", "London", "London", "London", "London", "Lon…
#> $ Sensor <chr> "Room 1", "Room 1", "Room 1", "Room 1", "Room 1", "Roo…
#> $ Date <dttm> 2024-01-01 00:00:00, 2024-01-01 00:15:00, 2024-01-01 …
#> $ Temp <dbl> 21.8, 21.8, 21.8, 21.7, 21.7, 21.7
#> $ RH <dbl> 36.8, 36.7, 36.6, 36.6, 36.5, 36.2
#> $ DewP <dbl> 6.383970, 6.344456, 6.304848, 6.216205, 6.176529, 6.05…
#> $ Abs <dbl> 7.052415, 7.033251, 7.014087, 6.973723, 6.954670, 6.89…
#> $ Mould <chr> "No mould", "No mould", "No mould", "No mould", "No mo…
#> $ PI <dbl> 45.25849, 45.38181, 45.50580, 46.07769, 46.20393, 46.5…
#> $ RH_if_2C_higher <dbl> 32.81971, 32.73052, 32.64134, 32.63838, 32.54920, 32.2…graph_TRH().mydata |>
mutate(DewPoint = calcDP(Temp, RH)) |>
graph_TRH() +
geom_line(aes(Date, DewPoint), col = "cyan3") + # add dew point
theme_bw()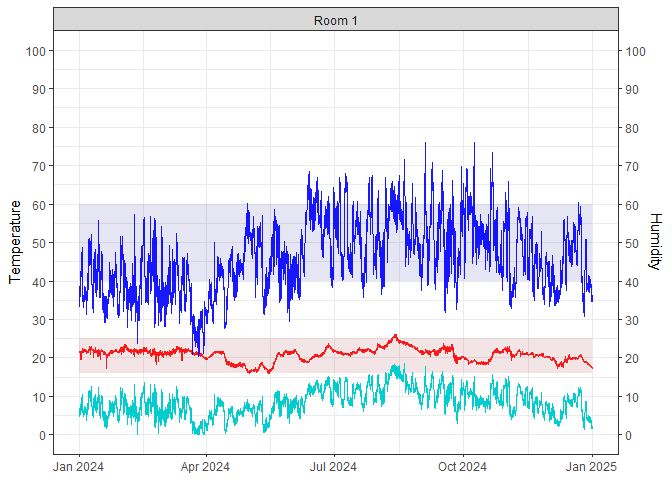
calcMould_Zeng() function and visualise it
alongside humidity data.mydata |>
mutate(Mould = calcMould_Zeng(Temp, RH)) |>
ggplot() +
geom_line(aes(Date, RH), col = "royalblue3") +
geom_line(aes(Date, Mould), col = "darkorchid", size = 1) +
labs(title = "Mould Growth Rate Limits",
subtitle = "Mould growth initiates when RH goes above threshold",
x = NULL, y = "Humidity (%)") +
facet_grid(~Sensor) +
theme_classic(base_size = 14)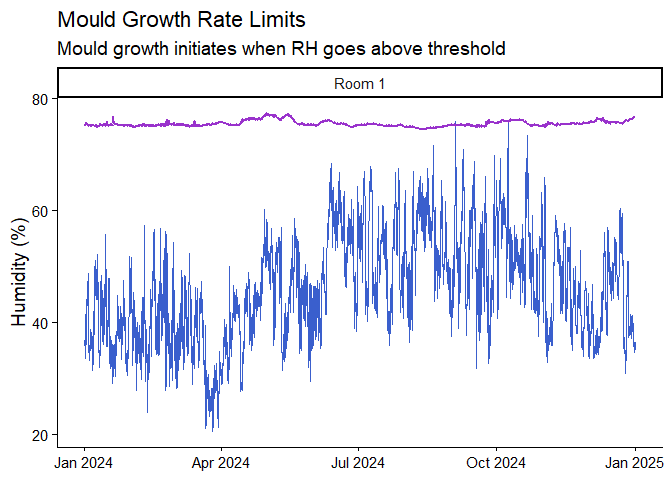
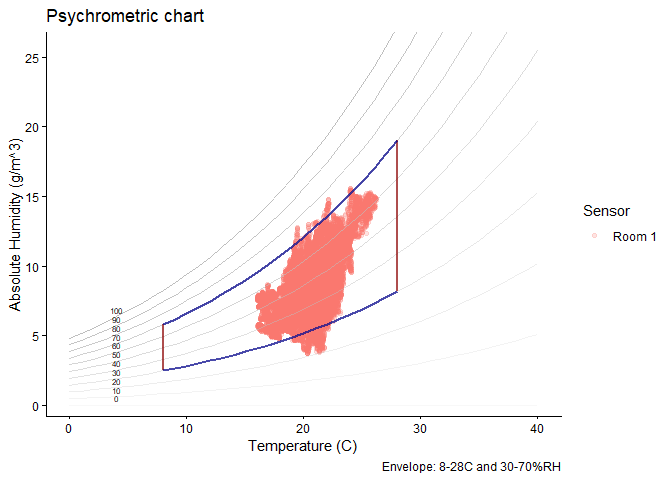
graph_psychrometric(). The example shows how a basic plot
can be customised; data transparency, temperature and humidity ranges,
and the y-axis function.# Customise
mydata |>
graph_psychrometric(
data_alpha = 0.2,
LowT = 8,
HighT = 28,
LowRH = 30,
HighRH = 70,
y_func = calcAH
) +
theme_classic() +
labs(title = "Psychrometric chart")